library(tidyverse)
library(survival)
library(ggsurvfit)
library(patchwork)In one course I give, students are asked to review a scientific article (a clinical trial in cancerology).
These same students have an R programming course with me later on, so I thought: what if the course content was to reproduce some of the figures in the study?
This would be interesting, educationally speaking, but the data from the clinical trial is obviously not available.
So here I am, trying to simulate data so that it gives results similar to those in the article.
The article (open-access), “Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer”, is about a phase 3 clinical trial testing the Progression-Free Survival of patients assigned to Trastuzumab Deruxtecan or another chemotherapy.
Scrape data from the article ♻️
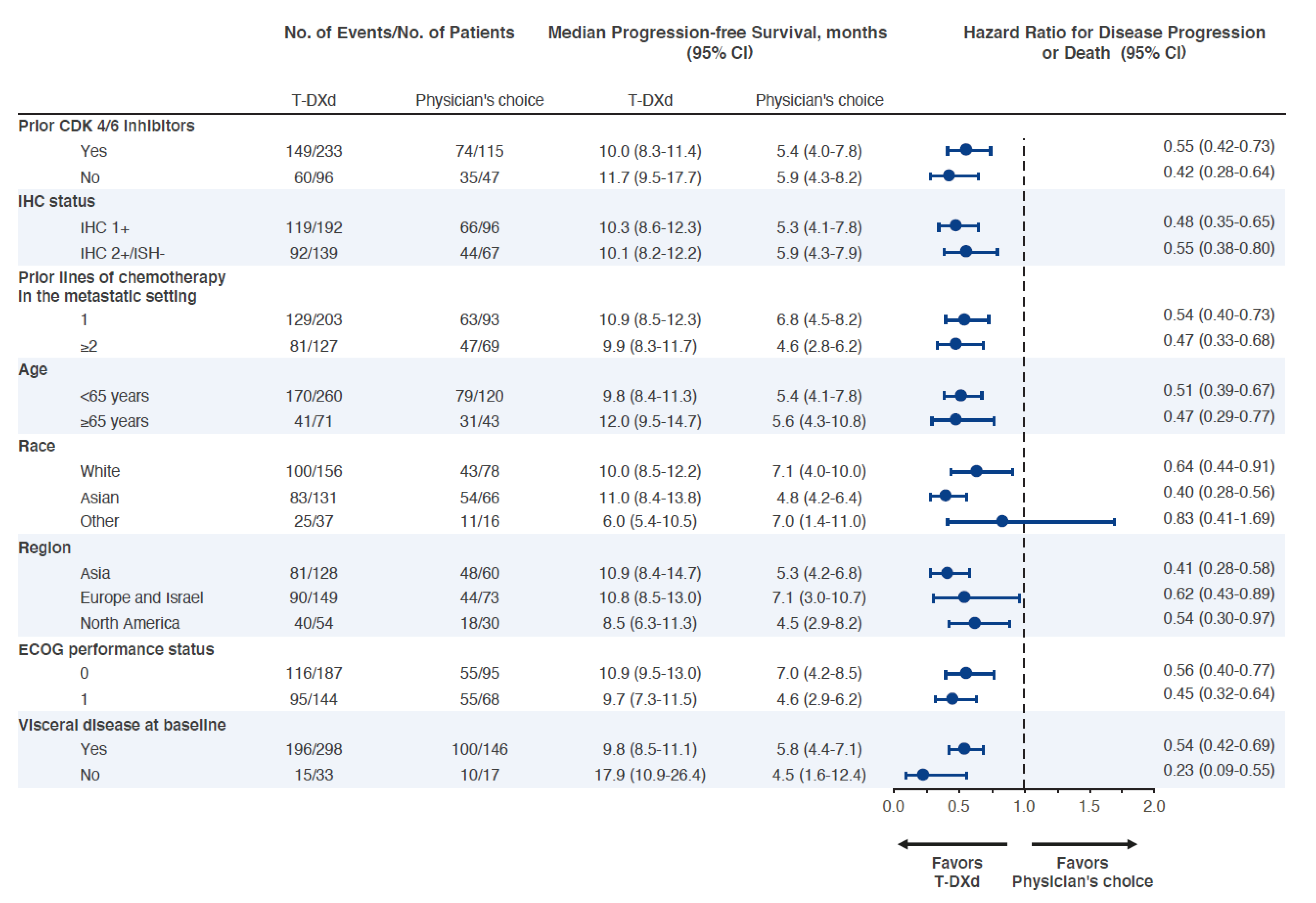
If I want to replicate the article, I will need some input data. The Figure S2 (Appendix, page 15) is perfect for that, and I think it will be the perfect figure for my course. Let’s scrape it!
Init
Here are the packages I will be using:
Baseline Data
First, I manually entered all the labels and sample sizes of all the covariates in sample() to generate a simulated population with the right baseline data.
As this implies some randomness, I encapsulate the process in the get_data() function and set the seed (more on that later).
Show the code for get_data()
#' Recreate article's baseline data
#' Data is from table S2 number of patients (`no_patients_tx` and `no_events_pc`)
get_data = function(){
f = function(lvl, nb) sample(rep(lvl, nb)) |> fct_relevel(lvl[1])
df0 = tibble(
id = factor(1:331),
arm = "T-DXd",
age = f(c("<65 years", "\U{2265}65 years"), c(260, 71)),
race = f(c("White", "Asian", "Other", NA), c(156, 131, 37, 7)),
region = f(c("Asia", "Europe and Israel", "North America"), c(128, 149, 54)),
ecog = f(c("0", "1"), c(187, 144)),
prior_cdki = f(c("Yes", "No", NA), c(233, 96, 2)),
prior_lines = f(c("1", "\U{2265}2", NA), c(203, 127, 1)),
ihc = f(c("IHC 1+", "IHC 2+/ISH-"), c(192, 139)),
visceral_bl = f(c("Yes", "No"), c(298, 33)),
)
df1 = tibble(
id = factor(331+c(1:163)),
arm = "Physician's choice",
age = f(c("<65 years", "\U{2265}65 years"), c(120, 43)),
race = f(c("White", "Asian", "Other", NA), c(78, 66, 16, 3)),
region = f(c("Asia", "Europe and Israel", "North America"), c(60, 73, 30)),
ecog = f(c("0", "1"), c(95, 68)),
prior_cdki = f(c("Yes", "No", NA), c(115, 47, 1)),
prior_lines = f(c("1", "\U{2265}2", NA), c(93, 69, 1)),
ihc = f(c("IHC 1+", "IHC 2+/ISH-"), c(96, 67)),
visceral_bl = f(c("Yes", "No"), c(146, 17)),
)
bind_rows(df0, df1) |>
mutate(
arm = fct_relevel(arm, "Physician's choice"),
prior_cdki = fct_relevel(prior_cdki, "Yes"),
ihc = fct_relevel(ihc, "IHC 1+"),
prior_lines = fct_relevel(prior_lines, "1"),
age = fct_relevel(age, "<65 years"),
race = fct_relevel(race, "White", "Asian"),
region = fct_relevel(region, "Asia", "Europe and Israel"),
ecog = fct_relevel(ecog, "0"),
visceral_bl = fct_relevel(visceral_bl, "Yes"),
) |>
crosstable::apply_labels(
prior_cdki = "Prior CDK 4/6 inhibitors", ihc = "IHC status",
prior_lines = "Prior lines of chemotherapy in the metastatic setting",
age = "Age", race = "Race", region = "Region", ecog = "ECOG performance status",
visceral_bl = "Visceral disease at baseline"
)
}set.seed(517)
data_baseline = get_data()
data_baselineCoefficients Data
Bored to death by the first step, I changed strategy and directly fed ChatGPT-4o with figure S2, asking for an R dataframe containing the data. The result was nearly perfect, but I used constructive::construct() to improve the readability.
Show the code for data_coef
data_figS2 = tibble(
variable = rep(
c(
"Prior CDK 4/6 inhibitors", "IHC status",
"Prior lines of chemotherapy in the metastatic setting", "Age", "Race",
"Region", "ECOG performance status", "Visceral disease at baseline"
),
rep(c(2L, 3L, 2L), c(4L, 2L, 2L))
),
variable_key = rep(
c("prior_cdki", "ihc", "prior_lines", "age", "race", "region", "ecog", "visceral_bl"),
rep(c(2L, 3L, 2L), c(4L, 2L, 2L))
),
level = c(
"Yes", "No", "IHC 1+", "IHC 2+/ISH-", "1", "\U{2265}2", "<65 years",
"\U{2265}65 years", "White", "Asian", "Other", "Asia", "Europe and Israel",
"North America", "0", "1", "Yes", "No"
),
no_events_tx = c(149, 60, 119, 92, 129, 81, 170, 41, 100, 33, 25, 81, 90, 36, 116, 80, 196, 15),
no_patients_tx = c(233, 95, 192, 139, 203, 127, 260, 51, 156, 54, 37, 128, 149, 48, 187, 146, 298, 33),
no_events_pc = c(74, 35, 46, 44, 63, 39, 79, 21, 45, 18, 11, 82, 94, 40, 55, 58, 108, 15),
no_patients_pc = c(115, 47, 65, 66, 103, 72, 121, 29, 102, 40, 16, 128, 147, 48, 87, 91, 122, 16),
median_pfs_tx = c(10, 11.7, 10.8, 10.1, 10.9, 9.3, 9.8, 9.4, 10.5, 11, 9.6, 10.1, 9.8, 10.2, 9.8, 10, 9.8, 10.1),
ci_pfs_tx = c(
"8.3-11.4", "9.6-13.1", "8.6-12.3", "8.2-12.2", "8.5-12.5", "8.1-12.0",
"8.9-11.3", "7.4-11.7", "9.5-12.2", "7.8-NA", "7.4-NA", "8.3-11.9",
"8.6-11.7", "8.4-NA", "8.8-11.1", "8.4-11.4", "9.8-11.1", "9.6-NA"
),
median_pfs_pc = c(5.4, 5.9, 5.8, 6, 6.4, 6.8, 5.4, 5.8, 6.5, 5.7, 4, 5.7, 5.8, 5.6, 5.3, 6.1, 5.3, 5.7),
ci_pfs_pc = c(
"4.0-7.8", "4.8-7.2", "4.3-7.7", "3.9-7.9", "4.8-8.2", "4.5-8.2", "4.4-7.8",
"4.1-10.4", "4.7-8.1", "3.9-10.6", "1.4-11.0", "4.3-7.8", "4.6-7.4",
"3.4-8.1", "4.4-7.9", "4.5-7.8", "5.1-6.4", "5.6-NA"
),
hr = c(
0.55, 0.42, 0.48, 0.55, 0.54, 0.47, 0.51, 0.47, 0.64, 0.4, 0.83, 0.41, 0.62,
0.54, 0.56, 0.45, 0.54, 0.23
),
ci_inf = c(
0.42, 0.28, 0.35, 0.38, 0.4, 0.33, 0.39, 0.29, 0.44, 0.28, 0.41, 0.28, 0.43,
0.3, 0.4, 0.32, 0.42, 0.09
),
ci_sup = c(
0.73, 0.64, 0.65, 0.8, 0.73, 0.68, 0.67, 0.77, 0.91, 0.56, 1.69, 0.58, 0.89,
0.97, 0.77, 0.64, 0.69, 0.55
),
)
data_coef = data_figS2 |>
select(variable_key, level, coef=hr, ci_inf, ci_sup) |>
mutate(
across(coef:ci_sup, log),
se = abs(ci_inf-ci_sup)/2,
coef_rel = coef-log(0.51)
)data_coefCheck ✔️
Let’s stop for a second and explore the baseline dataset that we generated.
Table 1
Did we reproduced the left side of Table 1 ?
Almost. There are differences, some most likely due to analytic reasons (e.g. missing data in race), and some that I don’t understand (e.g. prior lines of chemotherapy).
library(crosstable)
data_baseline |>
crosstable(c(region, race, ecog, prior_cdki, prior_lines), by=arm) |>
as_flextable(keep_id=TRUE)arm | ||
|---|---|---|
Physician's choice (N=163) | T-DXd (N=331) | |
Region (region) | ||
Asia | 60 (36.8%) | 128 (38.7%) |
Europe and Israel | 73 (44.8%) | 149 (45.0%) |
North America | 30 (18.4%) | 54 (16.3%) |
Race (race) | ||
White | 78 (48.8%) | 156 (48.1%) |
Asian | 66 (41.2%) | 131 (40.4%) |
Other | 16 (10.0%) | 37 (11.4%) |
NA | 3 | 7 |
ECOG performance status (ecog) | ||
0 | 95 (58.3%) | 187 (56.5%) |
1 | 68 (41.7%) | 144 (43.5%) |
Prior CDK 4/6 inhibitors (prior_cdki) | ||
Yes | 115 (71.0%) | 233 (70.8%) |
No | 47 (29.0%) | 96 (29.2%) |
NA | 1 | 2 |
Prior lines of chemotherapy in the metastatic setting (prior_lines) | ||
1 | 93 (57.4%) | 203 (61.5%) |
≥2 | 69 (42.6%) | 127 (38.5%) |
NA | 1 | 1 |
Internal correlation
Also, are the generated values correlated?
In a real setting, they should all be somewhat correlated (notably race and region), but if we want to simulate the data, correlation can cause multicollinearity that will get in our way.
Luckily (is it?), the inter-variable correlation is very low:
library(ggcorrplot)
model.matrix(~0+., data=data_baseline |> select(-id, -arm) ) |>
cor(use="pairwise.complete.obs") |>
ggcorrplot(show.diag=FALSE, type="lower", lab=TRUE, lab_size=2)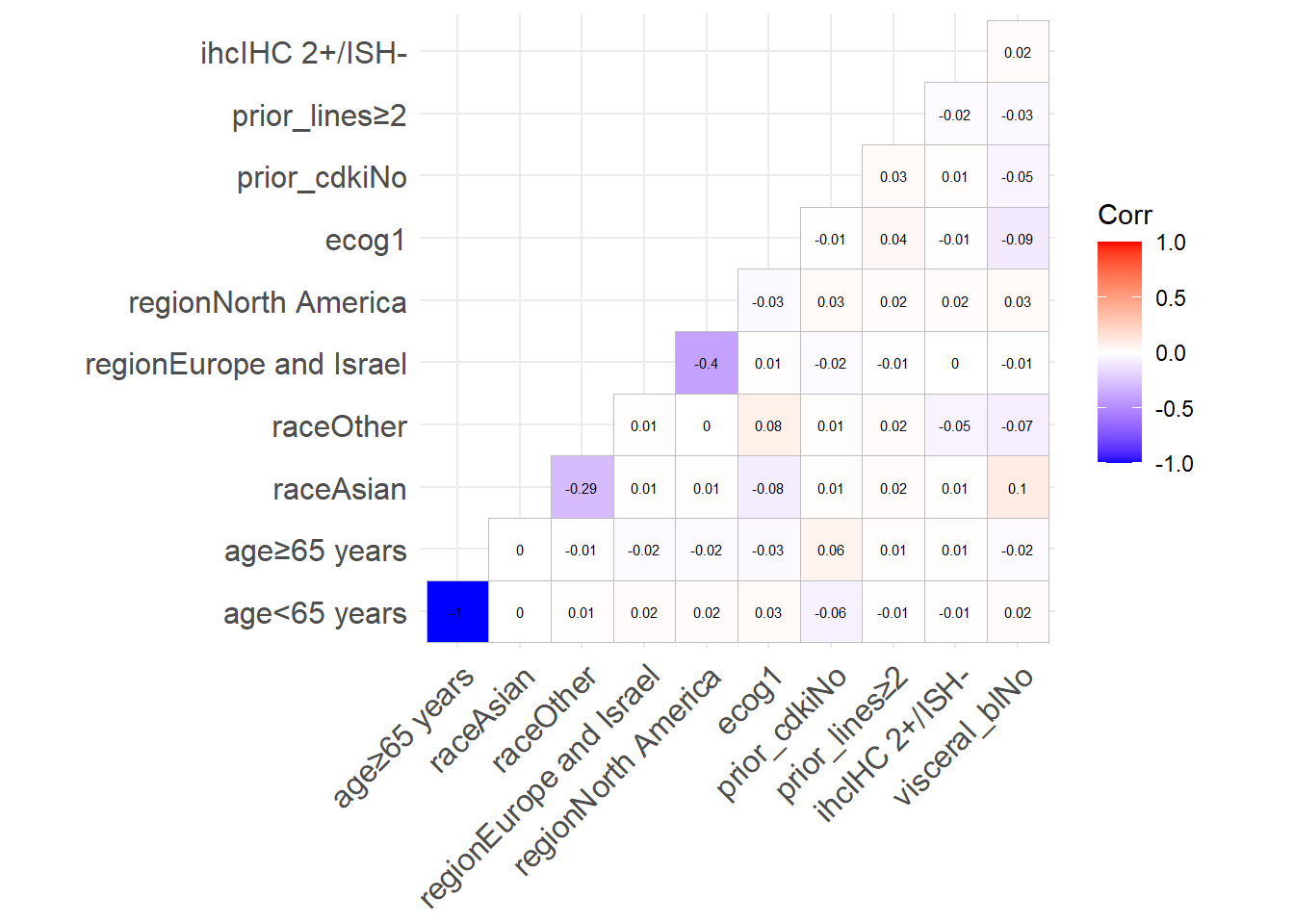
Calculation of times ⏱
OK, now we have baseline data and coefficients, we can generate survival times for our virtual patients.
To reproduce Figure S2, we need to simulate univariate interactions between all the covariates and the treatment arm.
Here, I used pivoting and joining to compute, for each patient, the sum of the coefficients corresponding to the values of their covariates.
I then use this sum, along with the coefficient for treatment arm, in the formula for a Weibull time.
I also generate a Weibull censorship time and set the end of study time to 20 months.
Similarly to the baseline data generation, this implies randomness, so I set the seed beforehand.
Show the code for add_surv_times()
# HR for PFS, 0.51; 95% CI, 0.40 to 0.64; P<0.001
beta_arm = log(0.51)
add_surv_times = function(data, a=2, b=0.55, eos=Inf){
data %>%
pivot_longer(-c(id, arm), names_to = "variable_key", values_to = "level") |>
left_join(data_coef, by = c("variable_key", "level")) |>
replace_na(list(coef=0, coef_rel=0)) |> # Impute missing data to 0
summarise(
coef_sum = sum(coef_rel),
.by=c(id)
) |>
right_join(data, by="id") |>
mutate(
arm_ttt = arm == "T-DXd",
logw = a + b * (log(rexp(n())) - (beta_arm+coef_sum)*arm_ttt),
time_event = exp(logw),
time_cens = rweibull(n=nrow(data_baseline), 5, 8), #5/8 cherry-picked with KM plots
time = pmin(time_event, time_cens, eos),
event = time<time_cens & time<eos,
) |>
mutate(
arm = fct_relevel(arm, "Physician's choice"),
prior_cdki = fct_relevel(prior_cdki, "Yes"),
ihc = fct_relevel(ihc, "IHC 1+"),
prior_lines = fct_relevel(prior_lines, "1"),
age = fct_relevel(age, "<65 years"),
race = fct_relevel(race, "White", "Asian"),
region = fct_relevel(region, "Asia", "Europe and Israel"),
ecog = fct_relevel(ecog, "0"),
visceral_bl = fct_relevel(visceral_bl, "Yes"),
)
}# set.seed(921)
set.seed(428)
data_surv = add_surv_times(data_baseline, eos=20)
data_survWe can also generate Kaplan-Meier curves to illustrate the data.
p1=ggsurvfit(survfit(Surv(time, event) ~ arm, data=data_surv))
p2=ggsurvfit(survfit(Surv(time, event) ~ arm+ecog, data=data_surv))
p1 + p2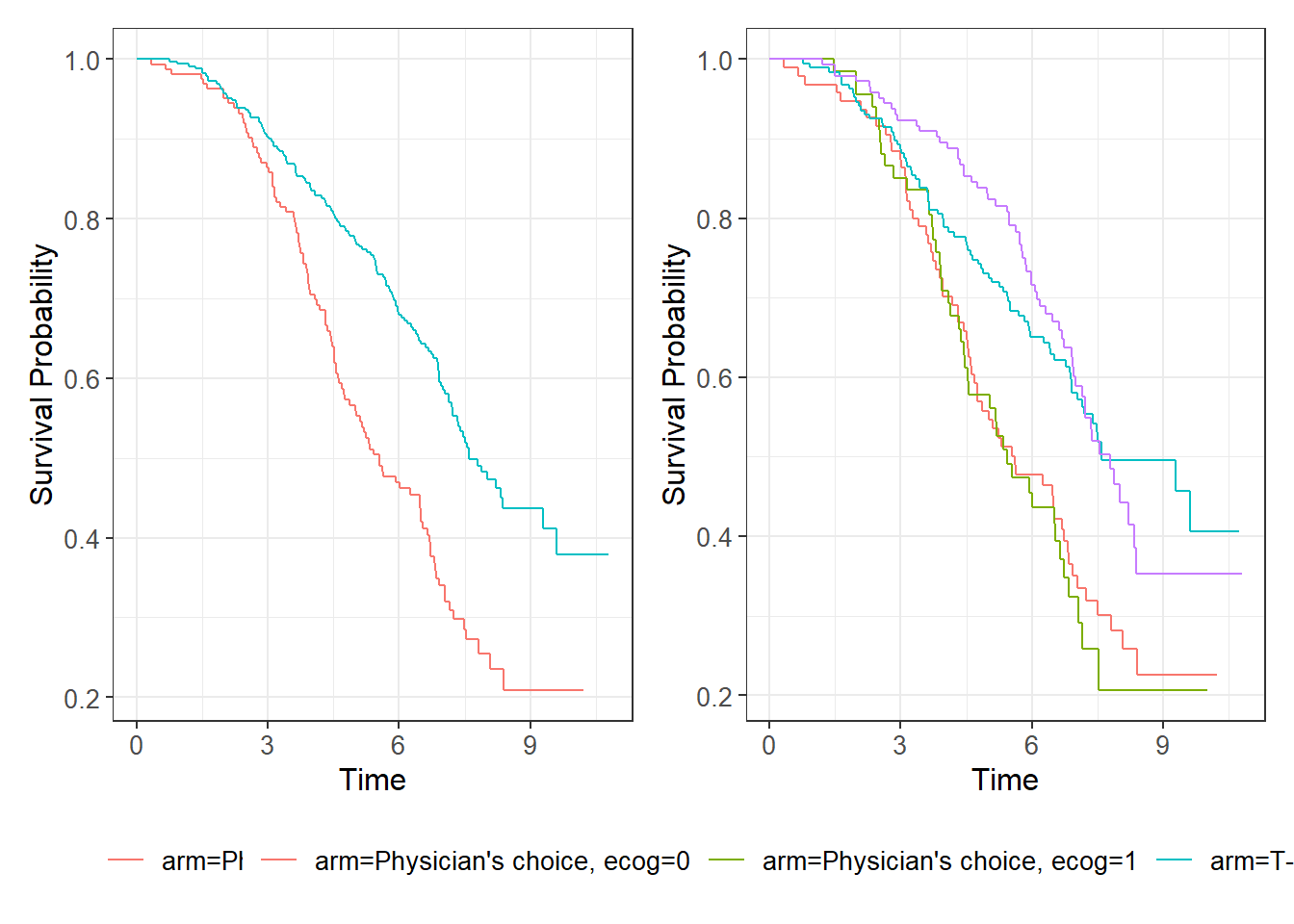
Mission completed 🚀
Hurray, we have simulated the dataset!

Now, can we reproduce the Figure S2 from our simulated data.
Show the code for coef_table()
coef_table = function(data, exp=TRUE){
variables = c("prior_cdki", "ihc", "prior_lines", "age", #figS2 order
"race", "region", "ecog", "visceral_bl")
variables |>
set_names() |>
map(~{
data |>
select(everything(), value=all_of(.x)) |>
filter(!is.na(value)) |>
summarise(
m = broom::tidy(coxph(Surv(time, event) ~ arm),
conf.int=TRUE, exponentiate=exp),
.by=value
) |>
unpack(m) |>
mutate(variable = .x, .before=0)
}) |>
list_rbind() |>
mutate(variable=as_factor(as.character(variable)),
value=fct_rev(as_factor(as.character(value)))) |>
arrange(variable, value)
}coef_surv = coef_table(data_surv)
coef_survp = coef_surv |>
ggplot(aes(y=fct_rev(value), x=estimate, xmin=conf.low, xmax=conf.high)) +
geom_point(shape="square", na.rm=TRUE) +
geom_errorbar(width = 0.6) +
geom_vline(xintercept=1, linetype="dashed", alpha=0.7) +
geom_vline(xintercept=exp(beta_arm), linetype="dotted", alpha=0.6) +
facet_grid(rows="variable", scale="free_y", ) +
labs(x="Hazard radio", y=NULL)
fig_s2 <- png::readPNG("fig_s2_mod.png", native=TRUE)
p + fig_s2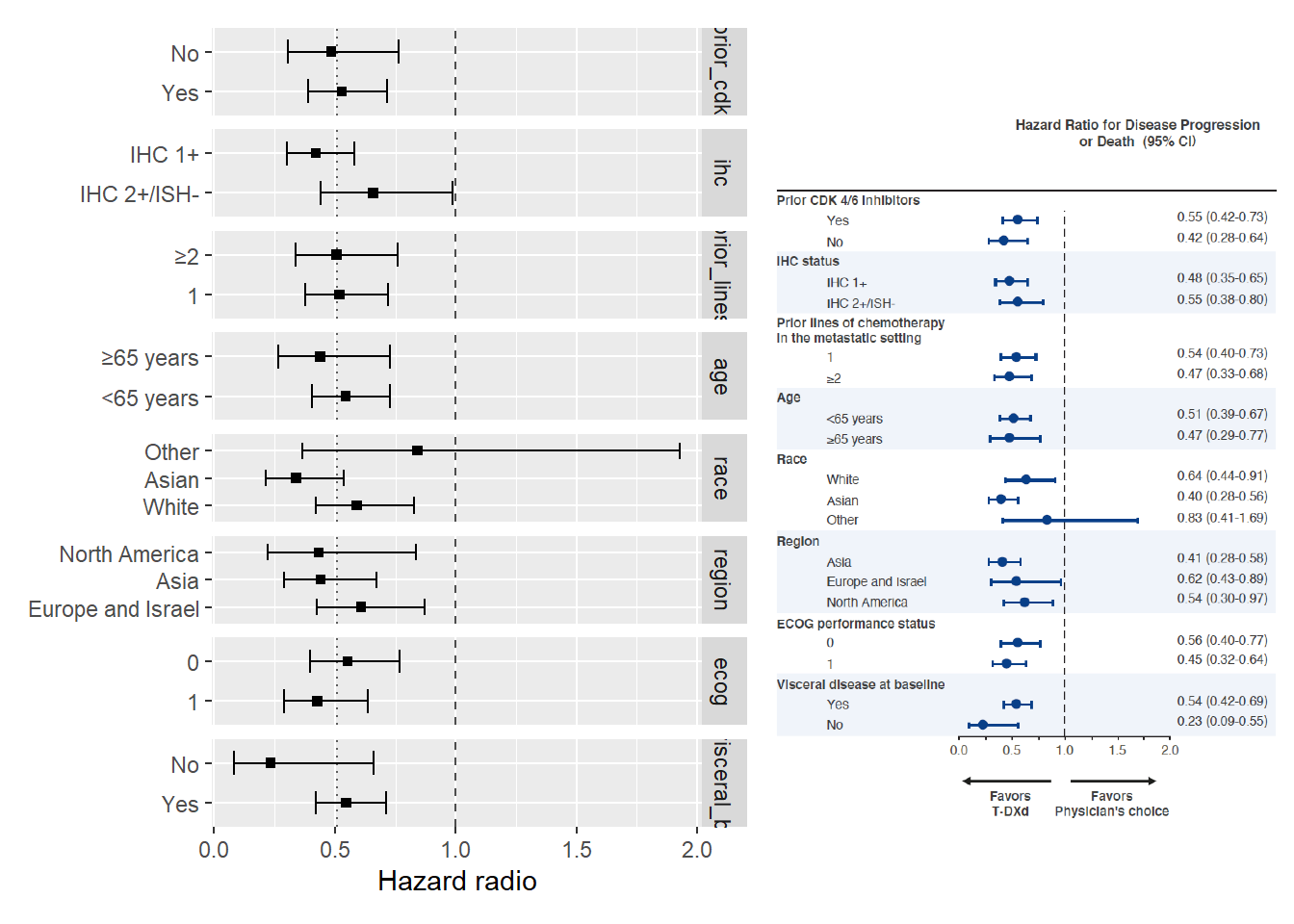
That’s not perfect, but it is close enough to play around during the course.
Post-production 🛠
Now that the proof of concept is done, the final touch will be to turn this dataset into something more real-lify, with dates instead of times.
library(lubridate)
accrual = as.Date(c("2018-12-27", "2021-12-31"))
destiny_breast04 = data_surv |>
mutate(date_enrolment = sort(sample(seq(ymd("2018-12-27"), ymd("2021-12-31"), by="day"), n())),
date_end_of_study = date_enrolment + time*365.24/12,
event_end_of_study = event,
) |>
select(id, arm, age:visceral_bl, date_enrolment,
date_end_of_study, event_end_of_study)
saveRDS(destiny_breast04, "destiny_breast04.rds")
destiny_breast04Now, the resulting dataset can be downloaded from destiny_breast04.rds.
Bonus: Seed optimizations 📈
Yeah, OK, I cheated a little…
In fact, I’ve been optimizing the seeds the whole time, so that the results looks the most like the original data.
Here is how I did it.
Seed optimization on correlation
Show the code for optim_correl_data()
optim_correl_data = function(seed, data_fun=get_data){
set.seed(seed)
d = data_fun() |> select(-id, -arm)
df_cor =
model.matrix(~0+., data=d) %>%
cor(use="pairwise.complete.obs")
df_cor[upper.tri(df_cor)] = 0
df_cor[abs(df_cor)==1] = 0
sum(abs(df_cor))
}
optim_correl_data = memoise::memoise(optim_correl_data, cache=cachem::cache_disk("cache"))cor_rslt = seq(1000) |>
set_names(~paste0("seed",.x)) |>
map_dbl(.progress=TRUE, ~{
optim_correl_data(.x)
})
cor_rslt[which.min(cor_rslt)] #seed517 = 1.978
#> seed517
#> 1.978512Seed optimization on coefs
Show the code for optim_coef_data()
optim_coef_data = function(seed, fun=add_surv_times, data=data_baseline){
set.seed(seed)
# d = data_surv
d = fun(data, eos=20)
x = coef_table(d, exp=FALSE) |>
rename(variable_key=variable, level=value) |>
left_join(data_coef, by=c("variable_key", "level")) |>
select(variable_key, level, coef, estimate) |>
mutate(delta = coef-estimate,
delta_rel = delta/coef)
x
}
optim_coef_data = memoise::memoise(optim_coef_data, cache=cachem::cache_disk("cache"))coef_rslt = seq(1000) |>
set_names(~paste0("seed",.x)) |>
map(.progress=TRUE, ~{
optim_coef_data(.x)
})
coef_rslt |> map_dbl(~sum(abs(.x$delta))) |> which.min() #seed921
#> seed921
#> 921
coef_rslt |> map_dbl(~sum(abs(.x$delta))) |> min() #1.278669
#> [1] 1.278669
coef_rslt |> map_dbl(~sum(abs(.x$delta_rel))) |> which.min() #seed428
#> seed428
#> 428Reuse
Citation
@online{chaltiel2025,
author = {Chaltiel, Dan},
title = {Replicate a Figure from an Article},
date = {2025-01-20},
url = {https://danchaltiel.github.io/blog/posts/2025-01_20_article_replication/},
langid = {en}
}